Challenge
Start of topic | Skip to actions
Workflow Proposals
Proposals should be submitted by Oct 24, 2008 For the Third Provenance Challenge the community will be using additional workflows beyond the brain imaging workflow used in the first two challenges. This page contains links to wiki pages describing the various candidate workflows that have been proposed by teams. To teams submitting workflows, please place a link under the Candidate Workflows section below describing your submission. Remember to include a paragraph about why the workflow is particularly novel. At the OpenProvenanceModelWorkshop, we had proposed a number of review criteria including- the workflows description should be expressed in english
- the workflows description should have figures
- component parts are available for download / source
- intermediate data made available for all components in the workflow
- the novelty of the workflow with respect to the challenge should be identified
Candidate Workflows
Two workflows from myExperiment (Manchester)
Here are two workflows from the myExperiment collection. In keeping with the myExp, er, experiment, we have created a single myExp "pack" to hold all info relevant to these. The pack belongs to Paul Fisher but should be visible by anyone: link to proposed Manchester workflows pack A brief description follows. I will add details to the pack if it turns out these are indeed interesting for the provenance challenge. The two bioinformatics flows are related to each other, having to do with- (1) finding all KEGG pathways that are associated to a specific region in the genome (a QTL, as these regions are known)
- (2) retrieving all PubMed? abstracts that are relevant to a KEGG pathway
- biomart
- pubmed
- KEGG
Trident Workflows from Astronomy (Pan-STARRS) and Oceanography (NEPTUNE)
Microsoft Research and Kno.e.sis Center, WSU
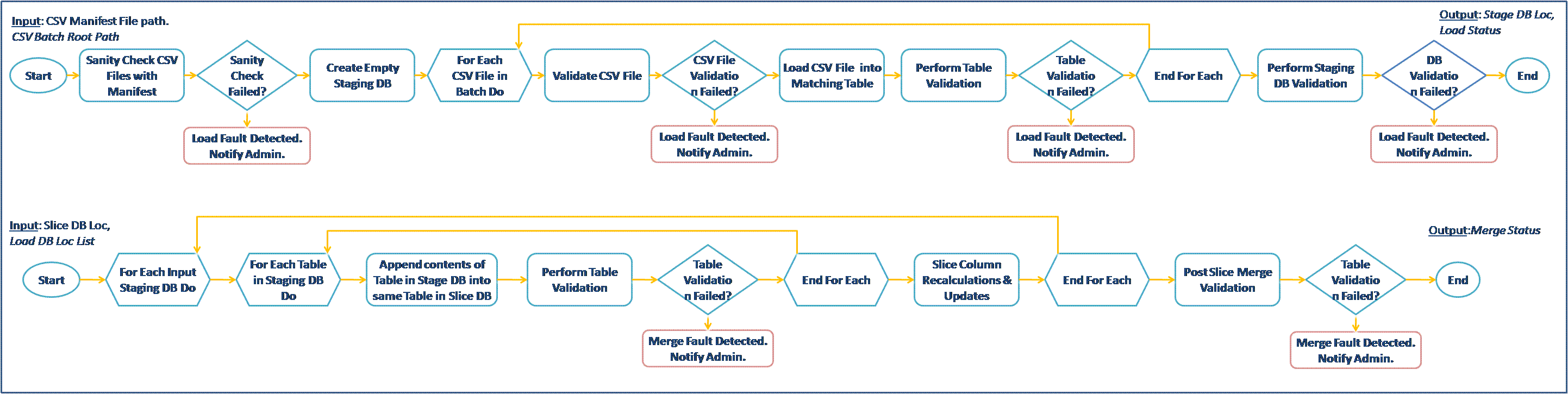
We propose two candidate workflows from the Neptune and Pan-STARRS eScience projects at Microsoft Research for the provenance challenge. We briefly describe the two scenarios here, the detailed description of the workflows, figures and the novelty of the scenarios for the challenge are in the (pdf) document at: MSR-WSU-Challenge3.pdf I. Oceanography Scenario (Neptune Project) Description: The Neptune project, led by the University of Washington (http://www.neptune.washington.edu/), is an ongoing initiative to create network of instruments widely distributed across, above, and below the seafloor in the northeast Pacific Ocean. We propose a simulated scenario involving collection of data by ocean buoys (containing a temperature sensor and an ocean current sensor) which is then sent as input to a scientific workflow for creation of a visualization chart as output. II. Astronomy Scenario (Pan-STARRS Project) Description: The Panoramic Survey Telescope & Rapid Response System (Pan-STARRS) will perform a detailed survey of the visible universe and build a time series of astronomical detections to track moving objects. Microsoft Research is working with University of Hawaii and Johns Hopkins University to create the infrastructure to manage large data generated by Pan-STARRS (~30TB/year) [Simmhan et. al, 2008]. We propose two workflows from the Pan-STARRS project for the provenance challenge. Resources for Pan-STARRS Workflows- The input CSV data files for the Load and Merge workflows along with schema for the databases will be provided. Alternatively, text files can be used to simulate the operation of the databases.
- Descriptions of validations to be performed will be provided. These are range checks on columns and simple astronomical checks that be easily implemented.
- A XOML representation of the Pan-STARRS workflows will be provided along with .NET libraries for the activities in the workflow. This will allow the workflows to be run using Windows .NET Workflow Runtime (Windows XP/Vista/Server 200X).
- MSR-WSU-Challenge3.pdf: Provenance Challenge 3 workflow entries from Pan-STARRS and NEPTUNE
Software build and testing (Luc and Paul)
I propose a simple workflow for building software systems. Specifically, I propose the ant file that compiles, builds and tests the OPM library in Java and Python. The ant build.xml file consists of a set of basic tasks (javac, xjc schema compiler, jar, testing) composed in two higher level targestsbuild.all and test.all.
The build.all target creates directories, compiles the OPM Schema, compiles all java files, produces a JAR file (and similarly for python, compiles the OPM schema, generating a python file). The test.all. will invoke some JUnit testing (still to be written sorry :-(.
The reference workflow is written with ant, but it could also be expressed as a Makefile, if it helps other teams such as PASS or ES3.
Capturing the provenance of the jar files and test results, will allow us to identify the tests that failed in a previous build, that passed in the latest, and correlate them to changed source files.
-- LucMoreau - 27 Oct 2008
Brain Imaging Workflows (Utah)
Julian Freire and Erik Anderson These workflows are use the VTK toolkit. Brain_uncolored_cortex.xml - This workflow represents the a 3D visualization of an extracted cortex. It contains sensor information colored by alpha power as computed by the stockwell transform at a single instant in time. These values reside at the sensor locations and nowhere else. Brain_3d_vis.xml - Using the above workflow, I take the scalar values from the sensors and use RBF interpolation to project them onto the cortical surface. Additionally, I also include the full 3D MRI to add contextual information to the visualization. Brain_plot.xml - This workflow represents a single sensor's information as a plot of the raw data as well as a plane representing the stockwell transform of the signal given at the specific sensor. This plane is oriented such that the y-axis is the alpha frequency band (8-12 Hz) and the x-axis is time (in samples, for the lifetime of the signal) Brain_final.xml - The culmination of the visualization representing the aggregation of the above workflows. This visualization combines Brain_plot with Brain_3d_vis and incorporates methods allowing them to interact with each other as the user picks various sensors. -- PaulGroth - 12 Nov 2008Reviews
to top
| I | Attachment  | Action | Size | Date | Who | Comment |
|---|---|---|---|---|---|---|
| | Pan-STARRS-LoadAndMergeWF.gif | manage | 77.7 K | 24 Oct 2008 - 05:46 | YogeshSimmhan | Pan-STARRS Load and Merge Workflows |
| | MSR-WSU-Challenge3.pdf | manage | 458.8 K | 24 Oct 2008 - 05:49 | YogeshSimmhan | Provenance Challenge 3 workflow entries from Pan-STARRS and NEPTUNE |
| | build.xml | manage | 3.0 K | 27 Oct 2008 - 10:48 | LucMoreau | build file for OPM library |
| | Brain_3d_vis.xml | manage | 43.8 K | 12 Nov 2008 - 19:02 | PaulGroth | Brain imaging vis-trails workflow |
| | Brain_final.xml | manage | 88.2 K | 12 Nov 2008 - 19:03 | PaulGroth | Brain imaging vis-trails workflow |
| | Brain_plot.xml | manage | 41.1 K | 12 Nov 2008 - 19:04 | PaulGroth | Brain imaging vis-trails workflow |
| | Brain_uncolored_cortex.xml | manage | 29.8 K | 12 Nov 2008 - 19:04 | PaulGroth | Brain imaging vis-trails workflow |
Edit | Attach image or document | Printable version | Raw text | More topic actions
Revisions: | r1.13 | > | r1.12 | > | r1.11 | Total page history | Backlinks
Revisions: | r1.13 | > | r1.12 | > | r1.11 | Total page history | Backlinks